View trace-plots of output from Bayesian segmentation model
Source:R/segmentation_helper_functions.R
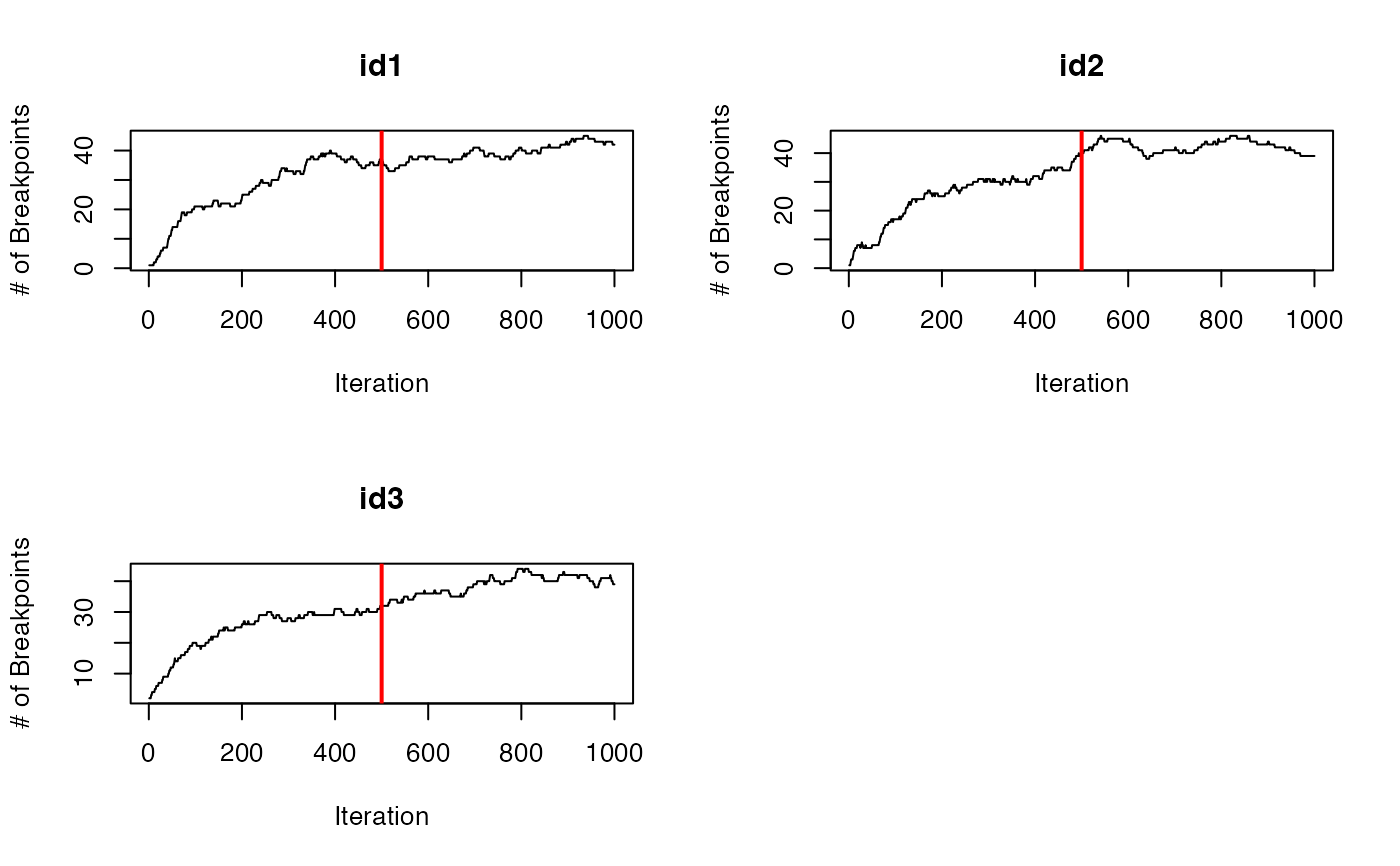
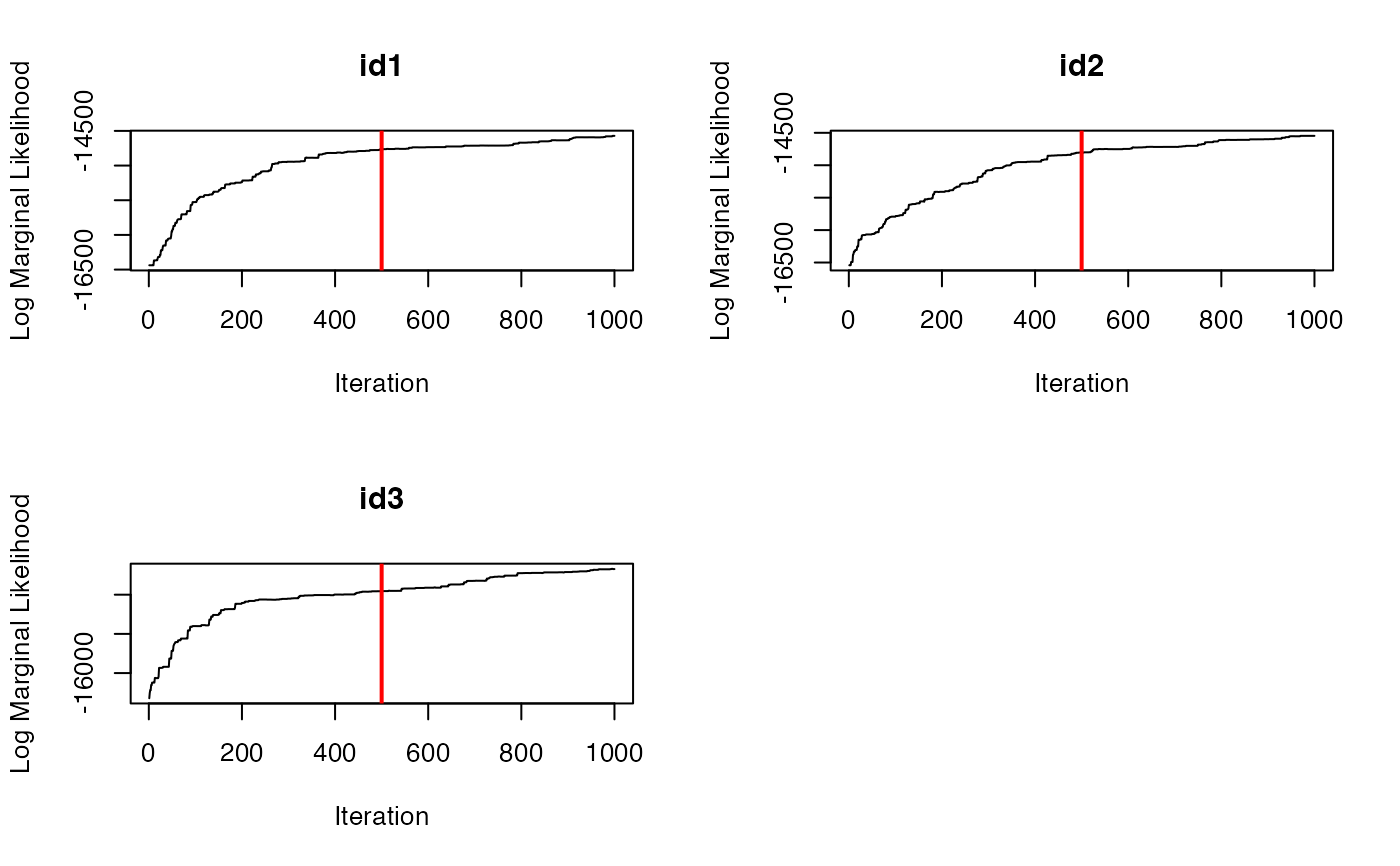
traceplot.RdVisualize trace-plots of the number of breakpoints estimated by the model as well as the log marginal likelihood (LML) for each animal ID.
traceplot(data, type)Arguments
- data
A list of model results that is returned as output from
segment_behavior.- type
character. The type of data that are being plotted from the Bayesian segmentation model results. Takes either 'nbrks' for the number of breakpoints or 'LML' for the log marginal likelihood.
Value
Trace-plots for the number of breakpoints or the log marginal likelihood are displayed for each of the animal IDs that were analyzed by the segmentation model.
Examples
# \donttest{
#load data
data(tracks.list)
#only retain id and discretized step length (SL) and turning angle (TA) columns
tracks.list2<- purrr::map(tracks.list,
subset,
select = c(id, SL, TA))
set.seed(1)
# Define model params
alpha<- 1
ngibbs<- 1000
nbins<- c(5,8)
future::plan(future::multisession, workers = 3) #run all MCMC chains in parallel
dat.res<- segment_behavior(data = tracks.list2, ngibbs = ngibbs, nbins = nbins,
alpha = alpha)
#> 4.379 sec elapsed
future::plan(future::sequential) #return to single core
#run function
traceplot(data = dat.res, type = "nbrks")
 traceplot(data = dat.res, type = "LML")
traceplot(data = dat.res, type = "LML")
 # }
# }